Rv2717c - conserved hypothetical protein
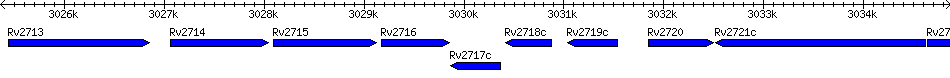
|
|
Protein Domains  |
| Gene Information | |
|---|---|
| Locus | Rv2717c |
| Symbol | |
| Gene Name | conserved hypothetical protein |
| Location | 3029867 - 3030361 (-) |
| Species | Mycobacterium tuberculosis H37Rv complete genome. |
| Length | Gene:495 bp Protein:165 aa |
| External Links | Tuberculist Target Gene Information String Protein-Protein Interactions STITCH Chemical-Protein Interactions Search Google Scholar |
Orthologs

|
| Orthogroup Number | 28135 |
| Related Genes | CE1476 MAP2833c MAV_3611 ML1006 Mmcs_0421 MSMEG_6574 MT2790 MUL_3360 Mvan_0449 nfa41140 SAV1365 | Transcriptional Regulation |
| Operons | View gene in operon browser |
| Regulatory Network | |
| Expression Correlation |
Genes with Correlated Expression Scatterplot of Gene Expression |