Rv3676 - transcriptional regulator, crp/fnr-family
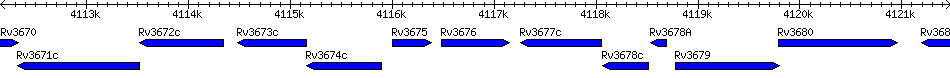
|
|
Protein Domains  |
| Gene Information | |
|---|---|
| Locus | Rv3676 |
| Symbol | |
| Gene Name | transcriptional regulator, crp/fnr-family |
| Location | 4116478 - 4117152 (+) |
| Species | Mycobacterium tuberculosis H37Rv complete genome. |
| Length | Gene:675 bp Protein:225 aa |
| External Links | Tuberculist Target Gene Information String Protein-Protein Interactions STITCH Chemical-Protein Interactions Search Google Scholar |
Orthologs

|
| Orthogroup Number | 33674 |
| Related Genes | Acel_1995 BL1216 CE0287 cg0350 DIP0303 jk1972 MAP0398c MAV_0453 Mkms_4904 ML2302 Mmcs_4818 MSMEG_6189 MT3777 MUL_4250 Mvan_5435 nfa3460 PPA0222 SAV4592 | Transcriptional Regulation |
| Operons | View gene in operon browser |
| Regulatory Network | |
| Expression Correlation |
Genes with Correlated Expression Scatterplot of Gene Expression |
| |||
| Proteins | |||
|---|---|---|---|
| Genomic Sequence | |||