Diversity Sequencing
TB Phylogeographic Diversity Sequencing Project
|
Different M. tuberculosis (TB) strains have varying virulence, immunogenicity, and drug resistance. However, currently sequenced T B genomes are thought to be responsible for only a minority of global TB burden. This project, led by Sebastien Gagneux and Peter Small in collaboration with the NIAID-funded Broad Genomic Sequencing Center for Infectious Disease, exploits recently revealed TB global population structure to re-sequence 25 rationally selected TB strains as representatives of the TB global diversity and identify sequence polymorphisms between these strains. Polymorphisms are searchable through the links below.
|
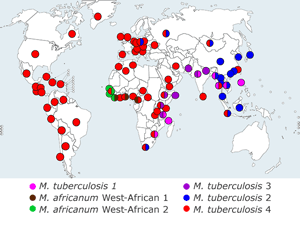
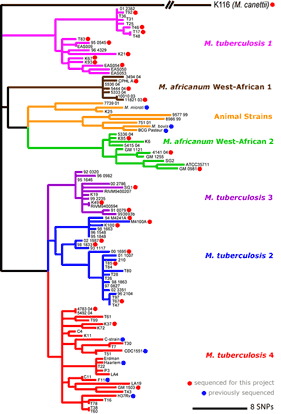
Maximum parsimony phylogenetic tree based on concatenates of 89 genes in 108 strains of M. tuberculosis complex (MTBC)(adapted from Hershberg et al 2008 PLoS Biology 6 (12):e311.). The 6 main lineages of human MTBC are indicated as in Gagneux & Small 2007 Lancet Infect Diseases 7(5):328-37. For comparison with other MTBC nomenclature see Comas et al 2009 PLoS ONE 12;4(11):e7815. Strains marked by a blue dot have full or partial whole-genome sequence available in the public domain. Strains marked by a red dot were sequenced (or are being sequenced) in this project. [Click image to enlarge] |
Download Data |
|
| Multiple Alignment of Diversity Sequencing Data | |